

To provide insight into the carbohydrate-protein interaction, GlycanInsight predicts the carbohydrate-binding pockets on a protein structure, analyzes the pocket characteristics, and suggests putative binding ligands of the protein.
Use the start page to submit your protein file.
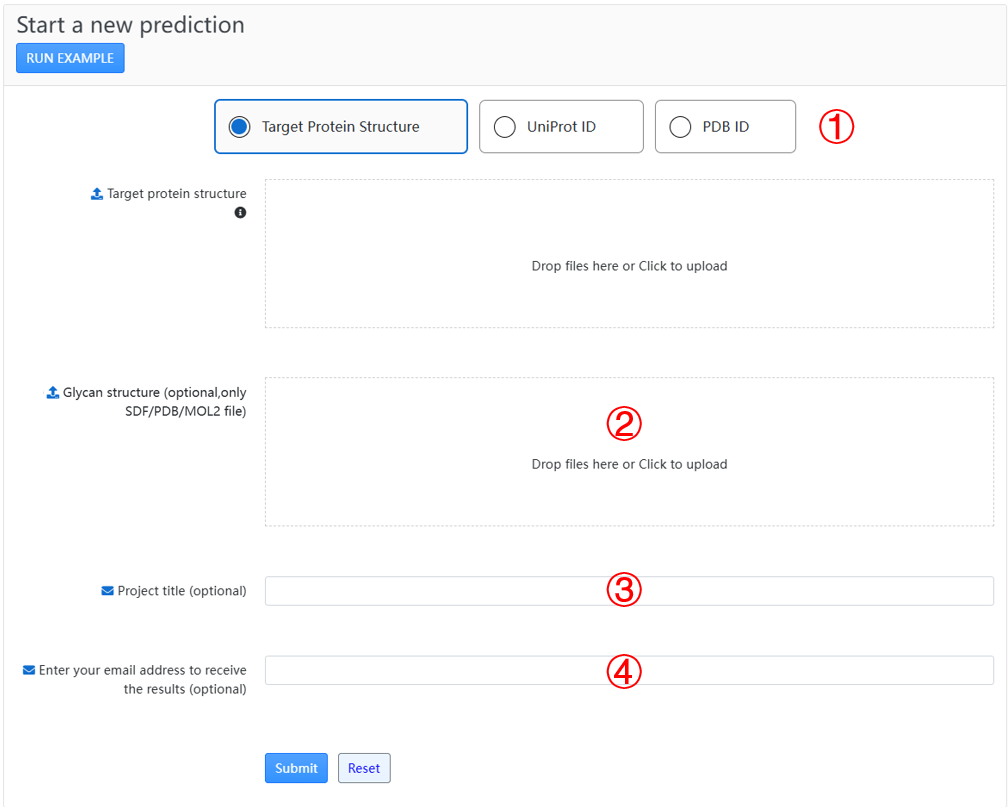
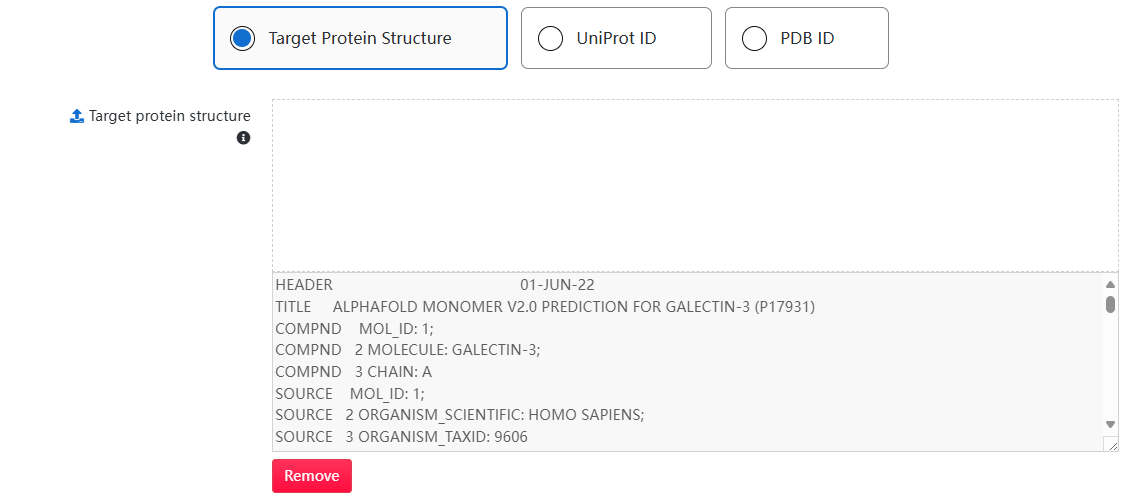
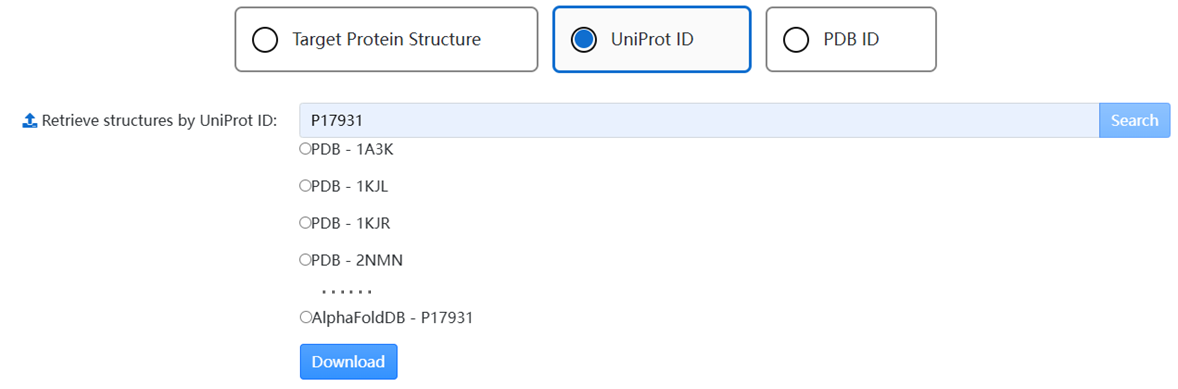
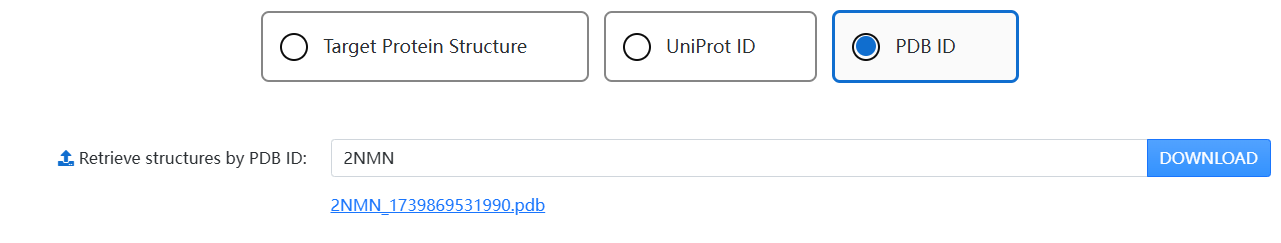
1. Use the tab to choose a way to obtain a structure:
2. You can specify a carbohydrate ligand for the query protein by uploading your ligand structure in SDF/PDB/MOL2 format. (not mandatory)
3. You can set a project title to name your experiment. (not mandatory)
4. You can use your email to receive your result page link and a compressed file containing all results. This field is not mandatory, but we recommend you use it to keep saved the address to your experiment. The link is the only way to access your online results.
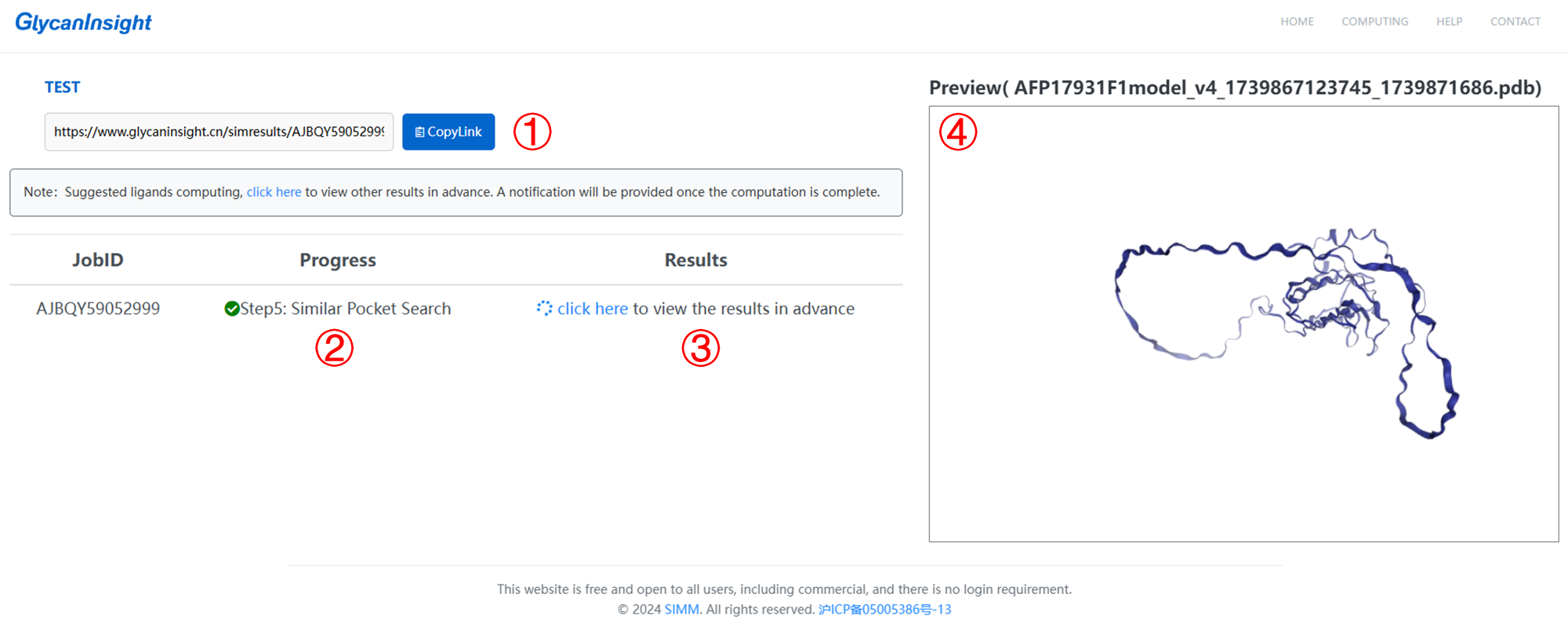
1. Here we provide a link to access the results or track the processing status of your task.
2. There are five steps: data preparation, glycan-binding pocket prediction, evolutionary conservation analysis, pocket cluster and putative ligands search.
3. Once step 4 (pocket cluster) is done, you can click to enter the result page in advance. A notification will be provided once the putative ligands search is complete.
4. Here you can preview the 3D structure of your submitted protein.
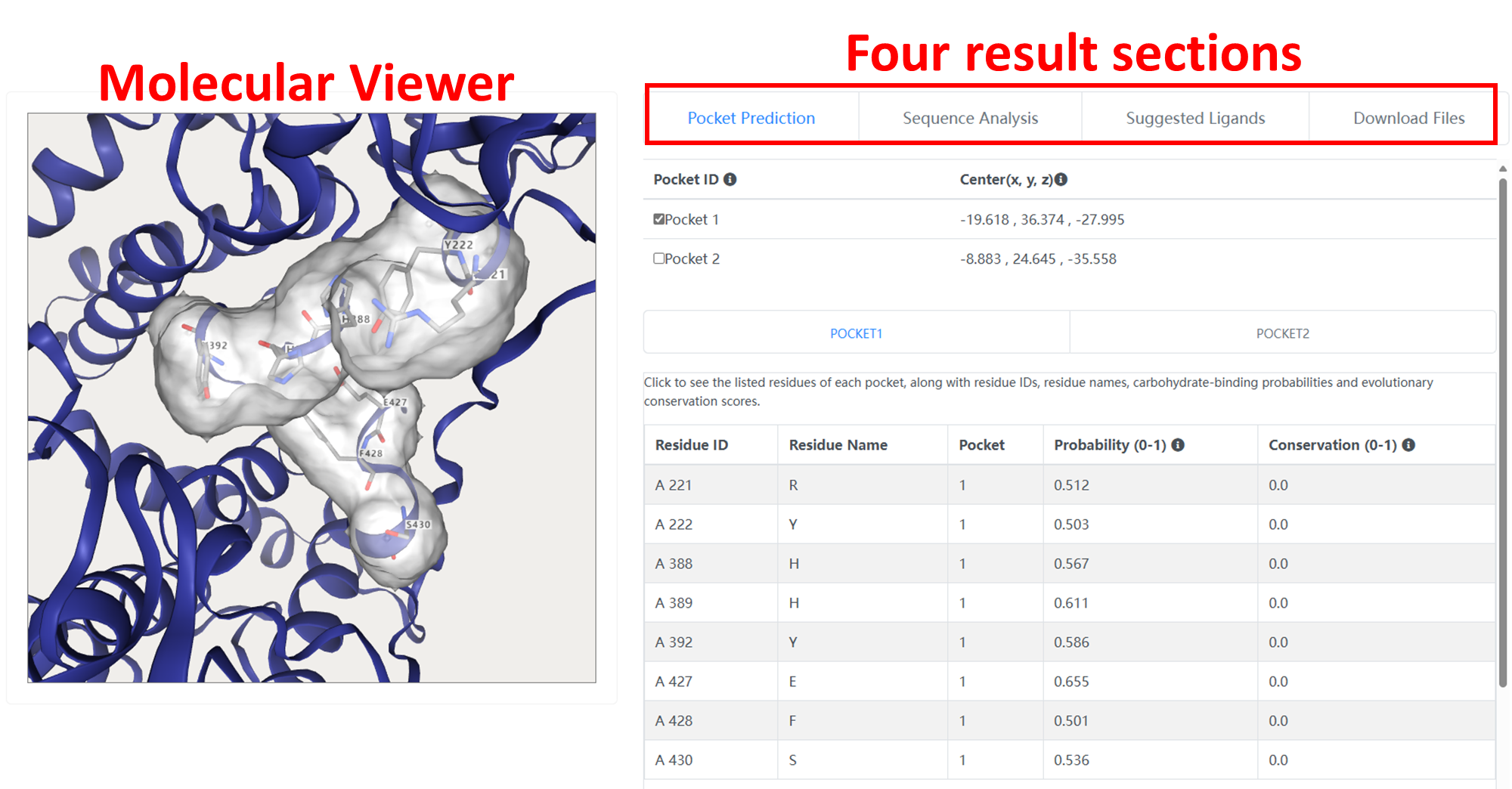
The result page of GlycanInsight is split into a molecular viewer and four result sections which can be chosen by tabs.
The visualization part contains an NGL molecular viewer that enables users to interactively inspect the predicted binding pockets within the protein structure.

The "Pocket Prediction" section shows detailed information of the predicted carbohydrate-binding pockets. This section is interactively coupled with the molecular viewer.
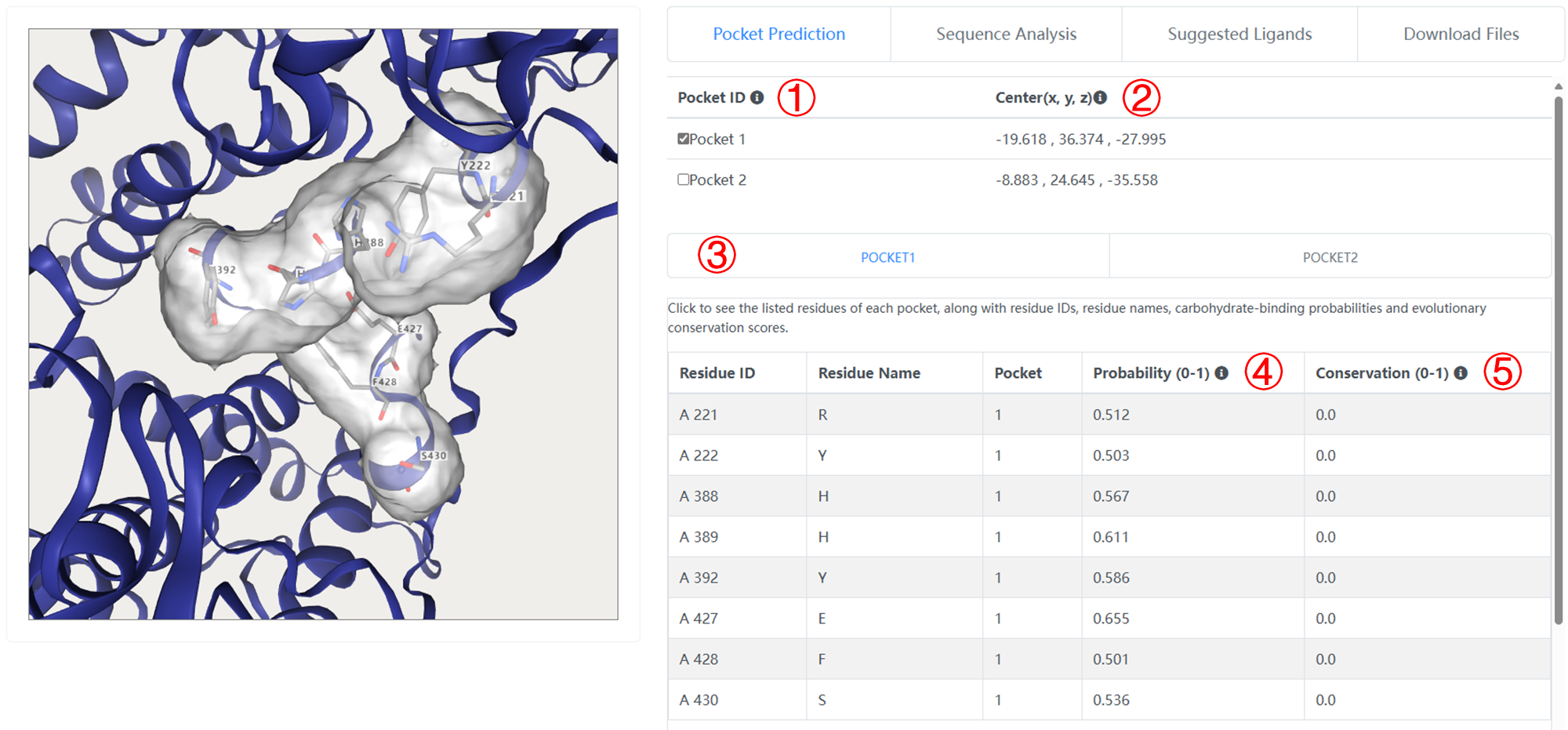
1. Click to inspect the pocket in molecular viewer window.
2. The center coordinate of each pocket.
3. Click to see the listed residues of each pocket, along with residue IDs, residue names, carbohydrate-binding probabilities and evolutionary conservation scores.
4. The carbohydrate-binding probability of per residue. GlycanInsight defines a residue with carbohydrate-binding probability larger than 0.5 to be a carbohydrate-binding pocket.
5. The evolutionary conservation score of per residue. The range of the conservation is between 0 and ~ 4 ( = log2(20) ) with higher values corresponding to higher conservation. GlycanInsight considers a site with conservation score larger than 0.5 to be a conserved one.
The "Sequence Analysis" section provides an easily interpretable view of the predicted carbohydrate-binding pockets.
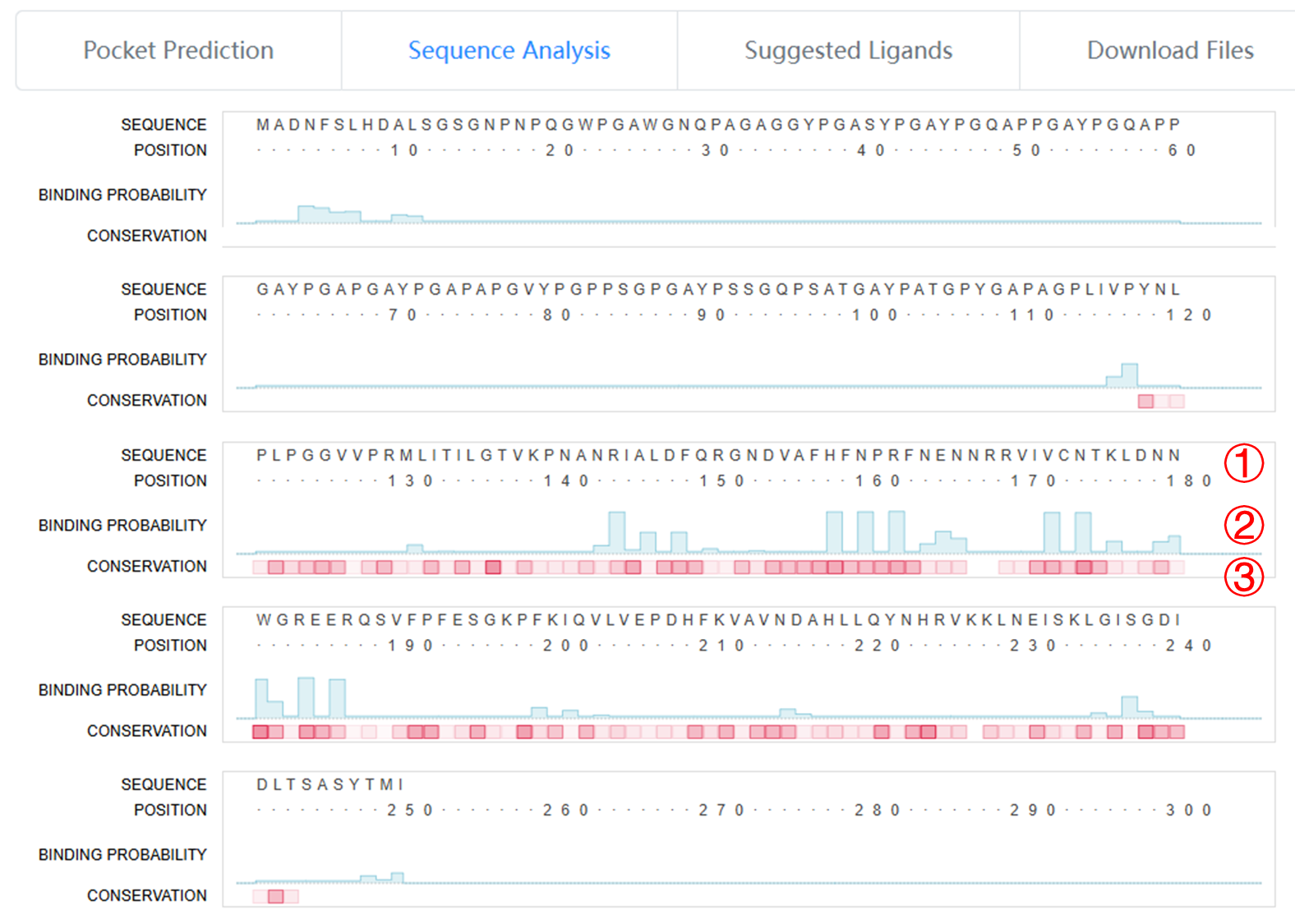
1. The amino acid sequence view of the submitted protein
2. The carbohydrate-binding probability presented in a column chart.
3. The conservation score indicated by the transparency level of red squares, with higher values corresponding to higher conservation.
The "Suggested Ligands" section lists putative binding ligands for the predicted carbohydrate-binding pockets, via comparing each predicted pocket against the known ligand-binding pocket. A table is provided for all ligand candidates, including:
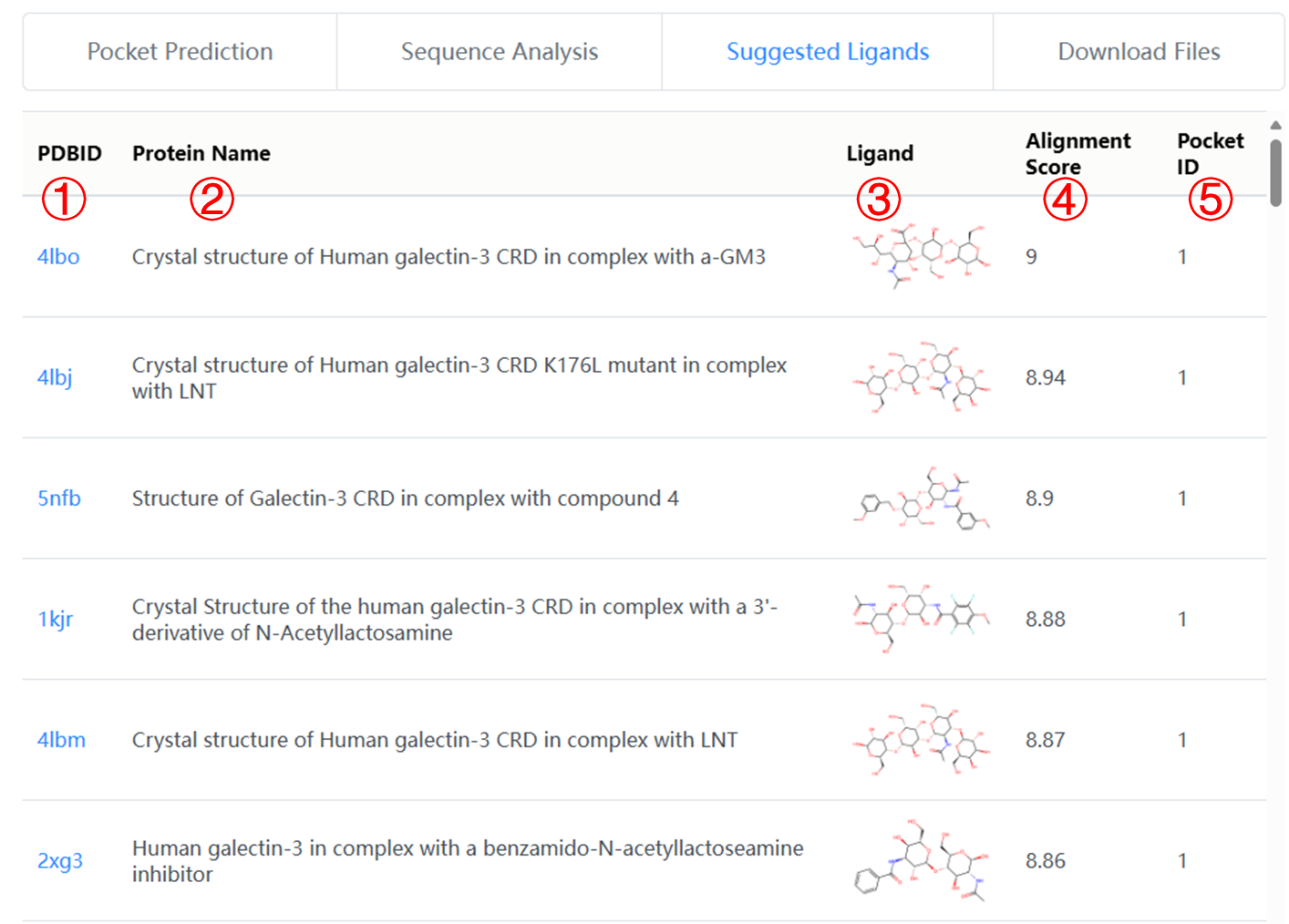
1. PDB ID of the protein containing similar known binding pocket.
2. Name of the protein containing similar known binding pocket.
3. Chemical structure diagram of the putative binding ligand.
4. Alignment scores between the predicted pocket and the known binding pocket.
5. Which pocket is calculated similar to the known binding pocket.
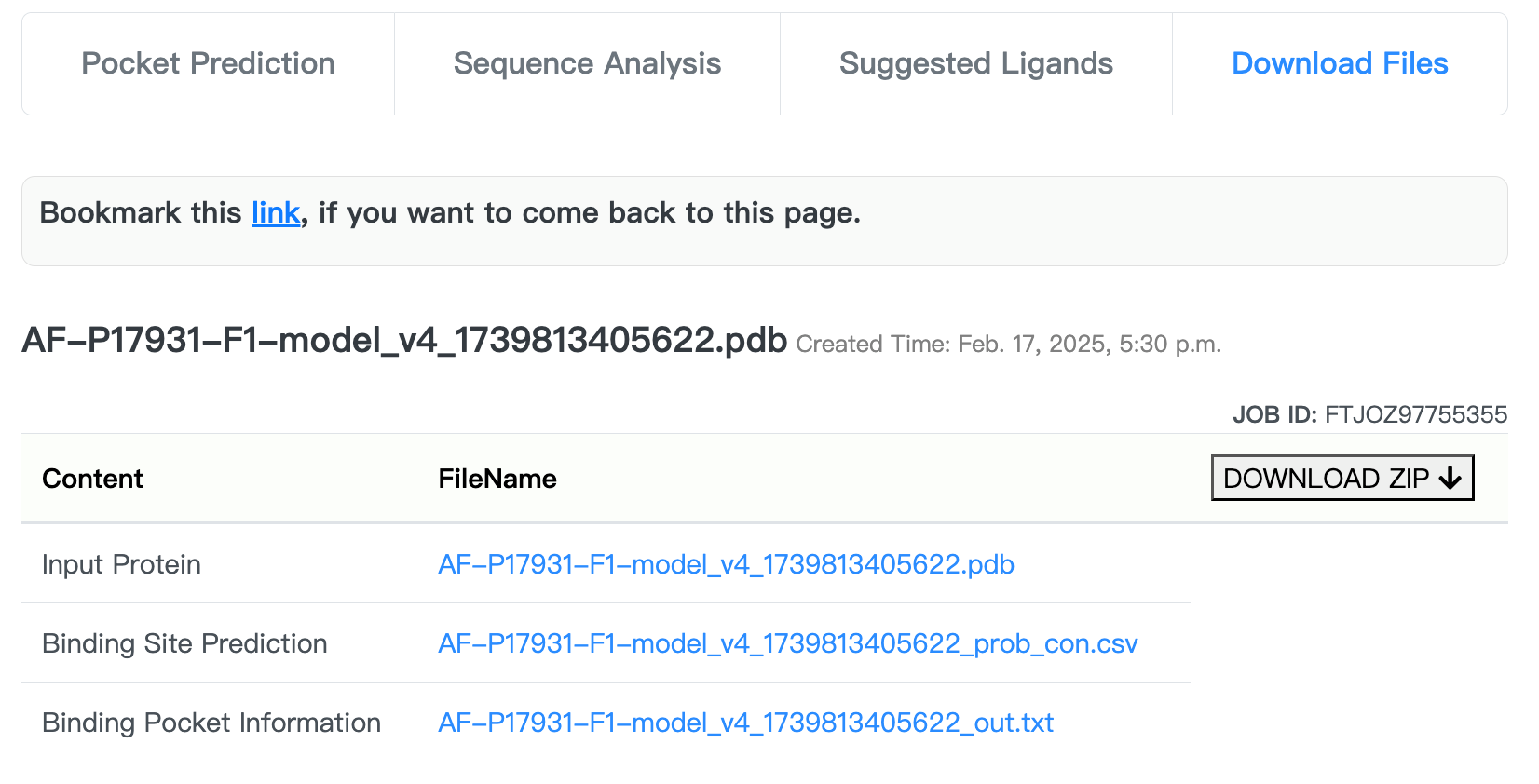
The "Download Files" section provides a buttom to download a compressed file containing protein and pocket PDB files, a PyMol visualization script and a list of prediction results.
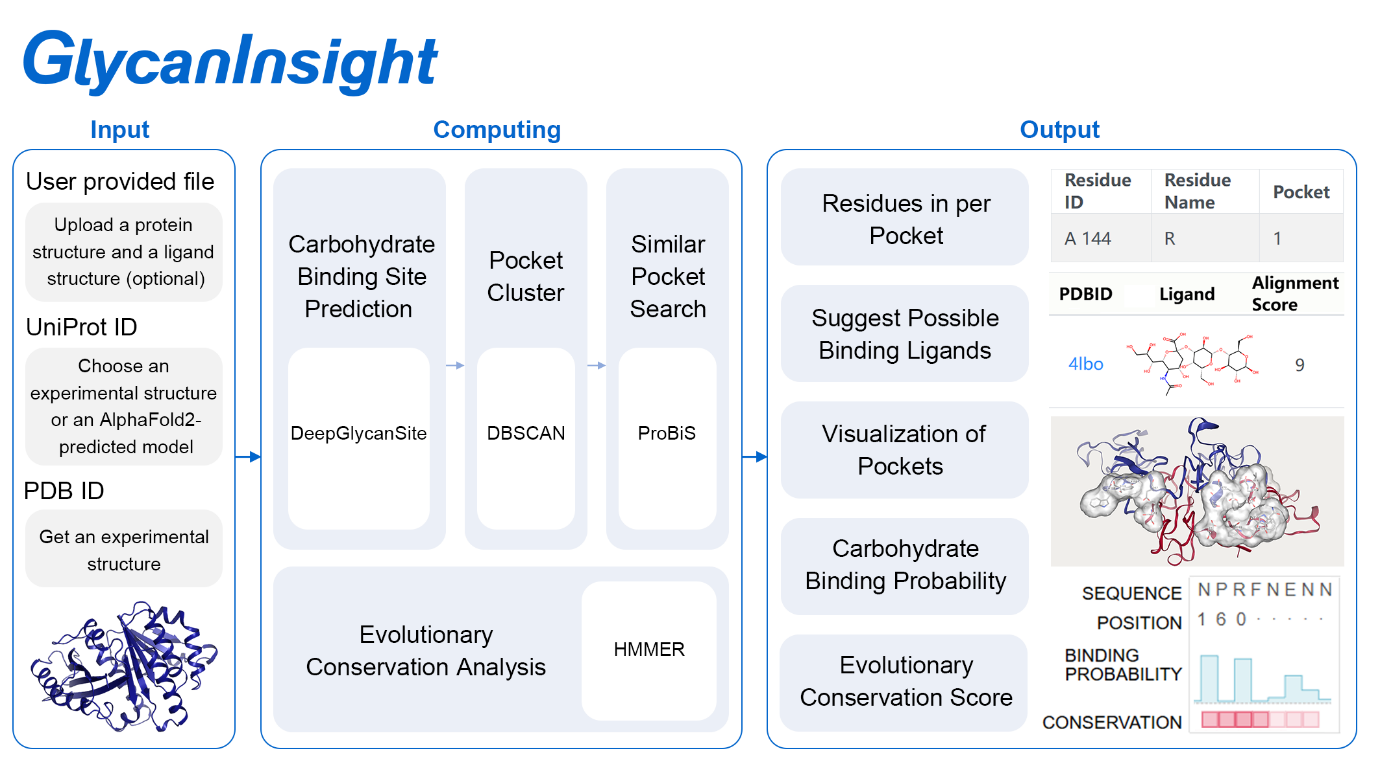
GlycanInsight used the DeepGlycanSite algorithm to predict the carbohydrate-binding probabilities of all protein residues. When the SDF file of a carbohydrate ligand is provided, GlycanInsight will use the ligand-version model (DeepGlycanSite+Ligand) to consider query ligand information. The residues with predicted probabilities larger than 0.5 are identified as carbohydrate-binding pockets. To obtain a more institutive understanding of the predicted carbohydrate-binding pockets on the query protein, GlycanInsight converts the residue-centric prediction result to a pocket-centric perspective using DBSCAN. By clustering the residues into regions that are potential pockets, we can estimate candidate bind ligands for each pocket. Assuming binding pockets bind to similar ligands, we use ProBiS to compare predicted pockets against the reported protein-ligand complexes. We list all reported ones with high alignment scores, indicating high similarity to the predicted pockets, and suggest their ligands as putative ligands for predicted pockets. Evolutionary conservation has been identified as a powerful indicator of functionally significant regions of proteins. To obtain more functional insights to the predicted carbohydrate-binding pockets, GlycanInsight also uses HMMER3 package to estimate their evolutionary conservation.